What's an unmixing matrix?
- olivertburton

- Aug 26
- 5 min read
In the last post, we looked at what the spectral mixing (sometimes called spillover) matrix looked like. Today, let's go through a bit about the unmixing matrix. I'm going to rely pretty heavily in parts on the unmixing-dependent spread paper by Peter Mage, Andrew Konecny and Florian Mair because this nicely illustrates what's going on in the unmixing. So, even though the examples and effect I'm going to show are different, the central idea comes from there.
To start, let's again look at the distinction between conventional and spectral flow cytometry. The basic thing that separates spectral from conventional flow is that we make more measurements than we have things we want to measure. We represent this information, these measurements across our fluorescent detector channels, as spectral profiles for the fluorophores we are trying to measure. I’m going to use this example of APC and PE-Cy5 to try to show a visual illustration of what’s going on. The APC, in red, has its highest emission when excited by the red laser, whereas PE-Cy5 is best excited by the yellow-green. They both have emission from all lasers, though.

We can represent these traces as heatmaps instead. This tends to be easier to read when there are lots of fluorophores. The spectral mixing matrix is rectangular—more detectors than fluorophores.

In conventional flow, we would have just one detector assigned to collect photons for PE-Cy5 and a second for APC. This would give us a square spillover matrix. This has much less information in it.

In conventional flow, we use a process called compensation to adjust the spillover signals out of the off-target channel and back into the assigned channel for that fluorophore. We want to achieve the result on the bottom, where we have a 1:1 relationship between signals in the detector and the desired fluorophore. We get this through using the compensation matrix and applying it to our collected data. The compensation matrix is simply in the inverse of the spillover matrix. It is what gives us this 1:0 matrix when multiplied by the spillover.

In spectral data, we have more information and don’t just do a simple removal of off-target signals. Instead, a common approach is to find a best fit using ordinary least-squares (OLS) modelling (if you're mathematically minded, take a trip to Wikipedia where you can read about the Moore-Penrose pseudoinverse and associated linear algebra to your heart's content). Anyway, we use this approach to generate an unmixing matrix rather than a compensation matrix. In the case of PE-Cy5 and APC, the OLS unmixing matrix looks like this:


What is this unmixing matrix telling us? If we look at the colour values, the areas of high intensity difference are around the peak of APC in the red and around the middle of the blue for PE-Cy5. These are the areas of biggest difference in emission between the two dyes. We do use the peak for PE-Cy5, but it is not as informative as other areas for discriminating the two. Note also that the UV is mostly ignored, which is consistent with there being minimal signal from either dye there.
When we add more fluorophores to our experiment, the individual signatures of the fluorophores, such as APC, do not change. The combination, the mixing matrix, is different, though.


And this means that the unmixing matrix also has to change. So, we will be using the information quite differently to assign the fluorophore values.

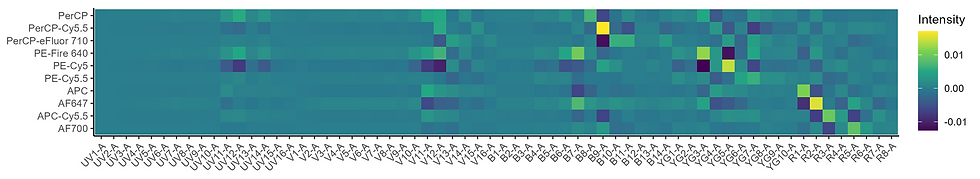
Compare the two unmixing matrices below. Both contain PE-Cy5 and APC. In the simple one, we are basically using all of the red signals to go to APC. This is very much not the case when we need to isolate more fluorophores with red emission. Separating these is more complicated. It’s pretty hard to look at the lower unmixing matrix and understand intuitively what’s going to happen to the data to achieve the unmixing. I’m going to suggest that the more difficult to interpret the unmixing matrix is, the more you will struggle with unmixing issues and data spread. Smaller errors will become more noticeable. See the unmixing-dependent spread paper for a more detailed explanation.


What can we do about this? We can improve our fluorophore combinations to achieve a cleaner spectral mixing matrix. In the example I’ve been showing you, there are many older fluorophores with repeating tandem elements. This gives us lots of repeated spillover patterns. Let’s look now at how swapping these to newer versions with less spillover can change the unmixing matrix.


Here are the full spectral mixing matrices. On the top, we have the one with older dyes. On the bottom, I’ve swapped out a few things, including changing APC to PE. Structurally, this looks pretty similar, but the bottom matrix is a bit higher contrast, which is what we want. This means there’s less off-peak emission. Note: this is based on what I've got lying around at the moment and isn't meant to be an "optimal" solution, just a better one.


The unmixing matrix for the set of messy older fluorophores looks a bit like a checkerboard—lots of ups and downs (note: this is all discussed thoroughly in the unmixing-dependent spread paper, see Figure SI 7 for this point). Notice how we’re having to rely on the UV emission, where there is minimal signal, to achieve unmixing. With the cleaner fluorophores, we can make better use of the blue, yellow-green and red, so we do not need to use the UV region, where there will be autofluorescence.

Crudely put: The easier the unmixing matrix is to interpret, the better the unmixing will work
Unmixing even an unstained sample is a bit problematic with this first set. With the cleaner fluorophores, the same fluorophore pair—AF647 and PE-Fire 640—is nicely centred around zero. So, the reliance on the UV for the first set is skewing the unmixing, even without any fluorophore signals present. This is because we aren't accounting for autofluorescence, and most of the signals in the UV are autofluorescence rather than from the fluorophores.

Anyway, that's all for today. Hopefully this has been helpful in understanding a bit about what's going on in the unmixing.
Given the central importance of the unmixing matrix for spectral flow cytometry data, it's unfortunate that it is not really accessible to most users. As far as I'm aware, this is not stored by any cytometer from any manufacturer in the FCS files. While the OLS unmixing matrix can be directly calculated from the spectral mixing matrix, the weighted least squares (WLS) unmixing matrix cannot. Furthermore, of the big three--Cytek Aurora, Sony ID7000 and BD S8/A8--only the BD instruments even provide the user with the spectral mixing matrix in the FCS files (stored in keyword SPILL).







Comments